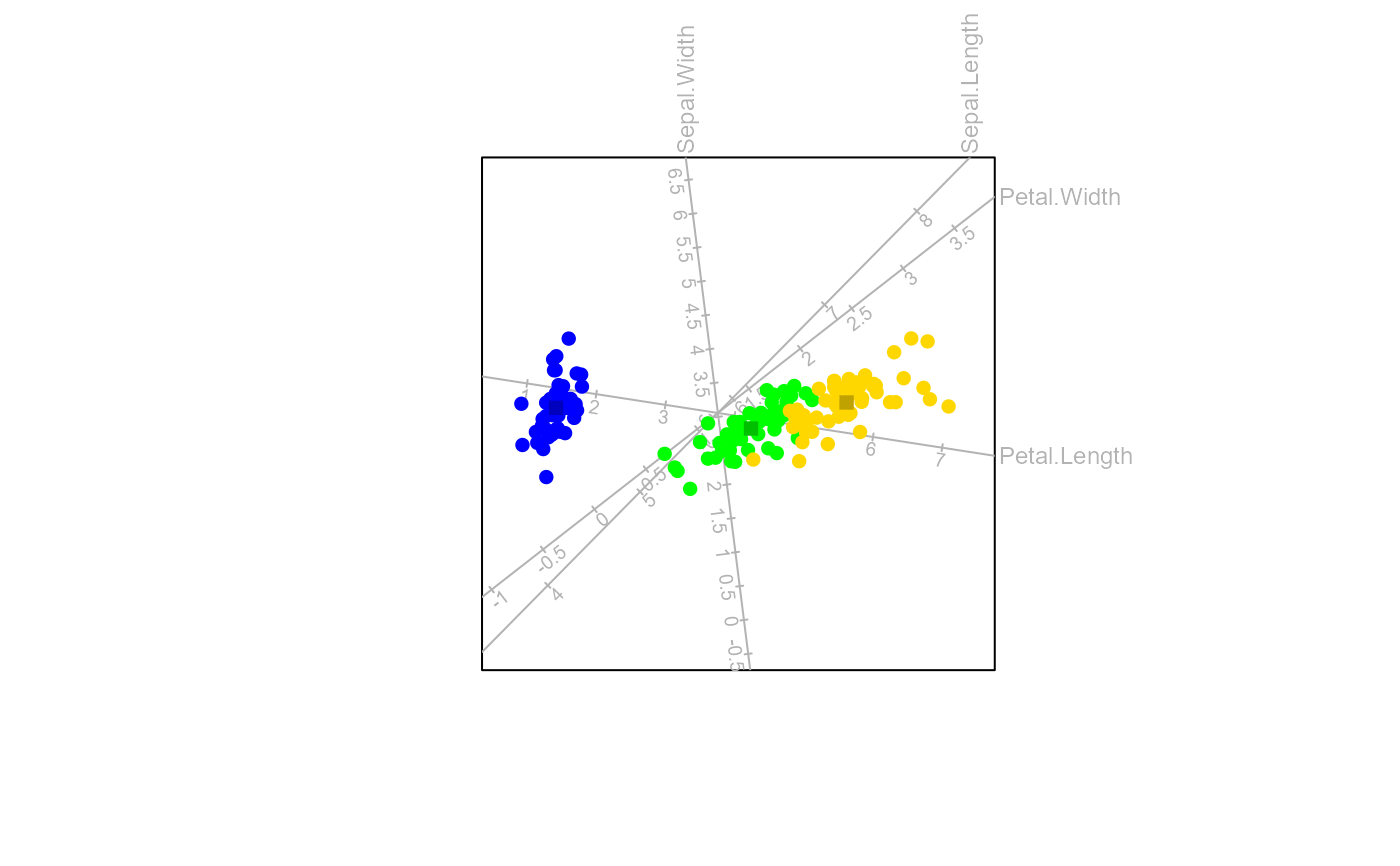
Use the Analysis of Distance (AoD) method to construct the biplot
AoD.RdThis function appends the biplot object with elements resulting from using the AoD method.
Arguments
- bp
an object of class
biplotobtained from preceding functionbiplot().- classes
a vector of the same length as the number of rows in the data matrix with the class indicator for the samples.
- dist.func
a character string indicating which distance function is used to compute the Euclidean embeddable distances between samples. One of
NULL(default) which computes the Euclidean distance or other functions that can be used for thedist()function.- dist.func.cat
a character string indicating which distance function is used to compute the Euclidean embeddable distances between samples. One of
NULL(default) which computes the extended matching coefficient or other functions.- dim.biplot
the dimension of the biplot. Only values
1,2and3are accepted, with default2.- e.vects
the vector indicating which eigenvectors (canonical variates) should be plotted in the biplot, with default
1:dim.biplot.- weighted
a character string indicating the weighting of the classes. One of "
unweighted" for each class to receive equal weighting or "weighted" for each class to receive their class sizes as weights.- show.class.means
a logical value indicating whether to plot the class means on the biplot.
- axes
a character string indicating the type of biplot axes to be used in the biplot. One of
"regression"or"splines".- ...
more arguments to
dist.func.