Class separation
Class_separation.Rmd
library(biplotEZ)
#> Welcome to biplotEZ!
#> This package is used to construct biplots
#> Run ?biplot or vignette() for more information
#>
#> Attaching package: 'biplotEZ'
#> The following object is masked from 'package:stats':
#>
#> biplotThis vignette deals with biplots for separating classes. Topics discussed are
- CVA (Canonical variate analysis) biplots
- AoD (Analysis of Distance) biplots
- Classification biplots
What is a CVA biplot
Consider a data matrix containing data on objects and variables. In addition, a vector contains information on class membership of each observation. Let indicate the total number of classes. CVA is closely related to linear discriminant anlaysis, in that the variables are transformed to new variables, called canonical variates, such that the classes are optimally separated in the canonical space. By optimally separated, we mean maximising the between class variance, relative to the within class variance. This can be formulated as follows:
Let be an indicator matrix with unless observation belongs to class and then . The matrix is a diagonal matrix containing the number of observations per class on the diagonal. We can form the matrix of class means . With the usual analysis of variance the total variance can be decomposed into a between class variance and within class variance:
The default choice for the centring matrix leads to the simplification
Other options are and . To find the canonical variates we want to maximise the ratio
subject to . It can be shown that this leads to the following equivalent eigen equations:
with and .
Since the matrix is symmetric and positive semi-definite the eigenvalues in the matrix are positive and ordered. The rank of so that only the first eigenvalues are non-zero. We form the canonical variates with the transformation
To construct a 2D biplot, we plot the first two canonical variates where . We add the individual sample points with the same transformation
where Interpolation of a new sample follows as . Using the inverse transformation , all the points that will predict for variable will have the form
where is a vector of zeros with a one in position . All the points in the 2D biplot that predict the value will have
defining the prediction line as
Writing the construction of biplot axes is similar to the discussion in the biplotEZ vignette for PCA biplots. The direction of the axis is given by . To find the intersection of the prediction line with we note that where is the length of the orthogonal projection of on .
Since is along we can write and all points on the prediction line project on the same point . We solve for from
If we select ‘nice’ scale markers for variable , then and positions of these scale markers on are given by with
with
The function CVA()
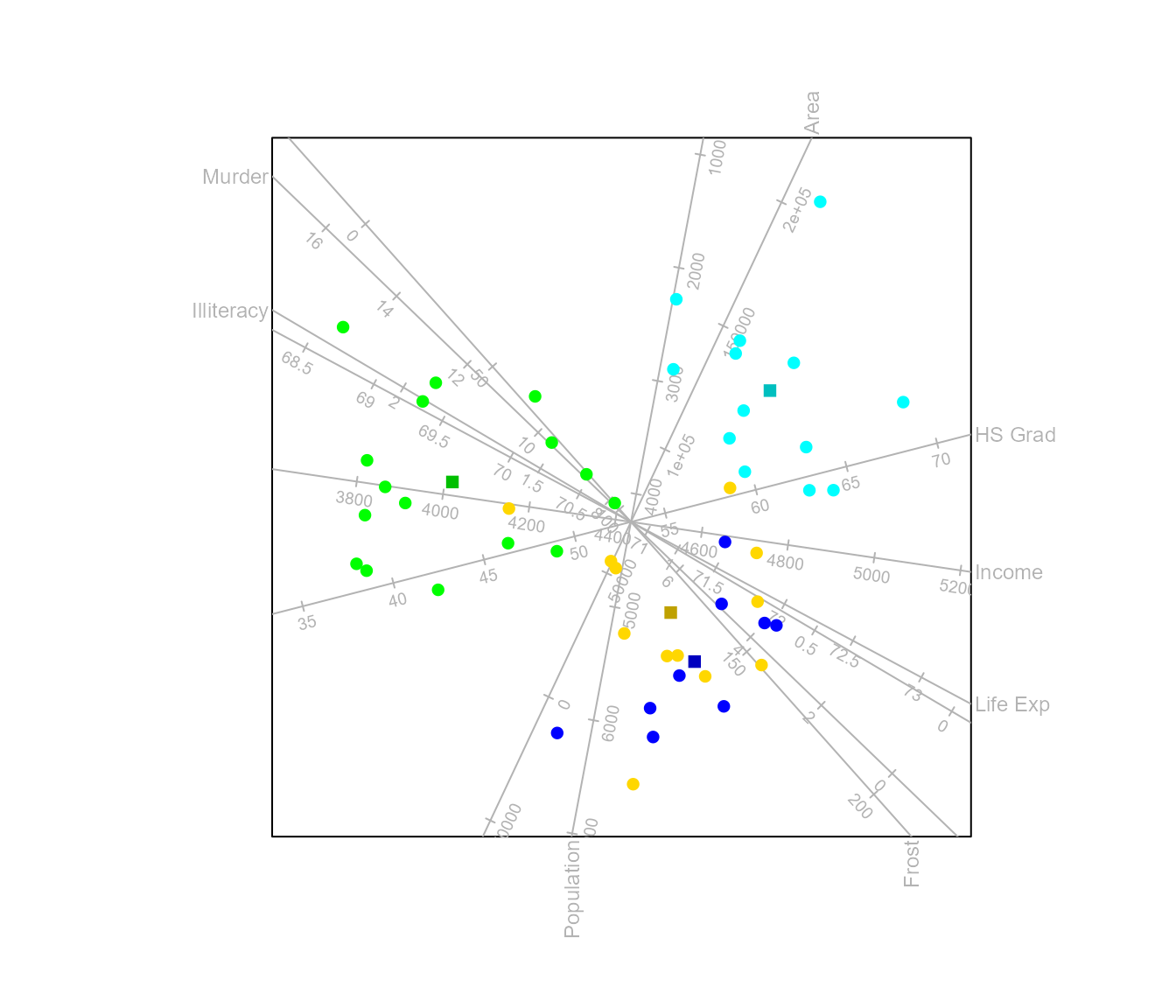
To obtain a CVA biplot of the state.x77 data set,
optimally separating the classes according to state.region
we call
Fitting -bags to the classes makes it easier to compare class overlap and separation. For a detailed discussion on -bags, see the biplotEZ vignette.
biplot(state.x77) |> CVA(classes = state.region) |> alpha.bags() |>
legend.type (bags = TRUE) |>
plot()
#> Computing 0.95 -bag for Northeast
#> Computing 0.95 -bag for South
#> Computing 0.95 -bag for North Central
#> Computing 0.95 -bag for West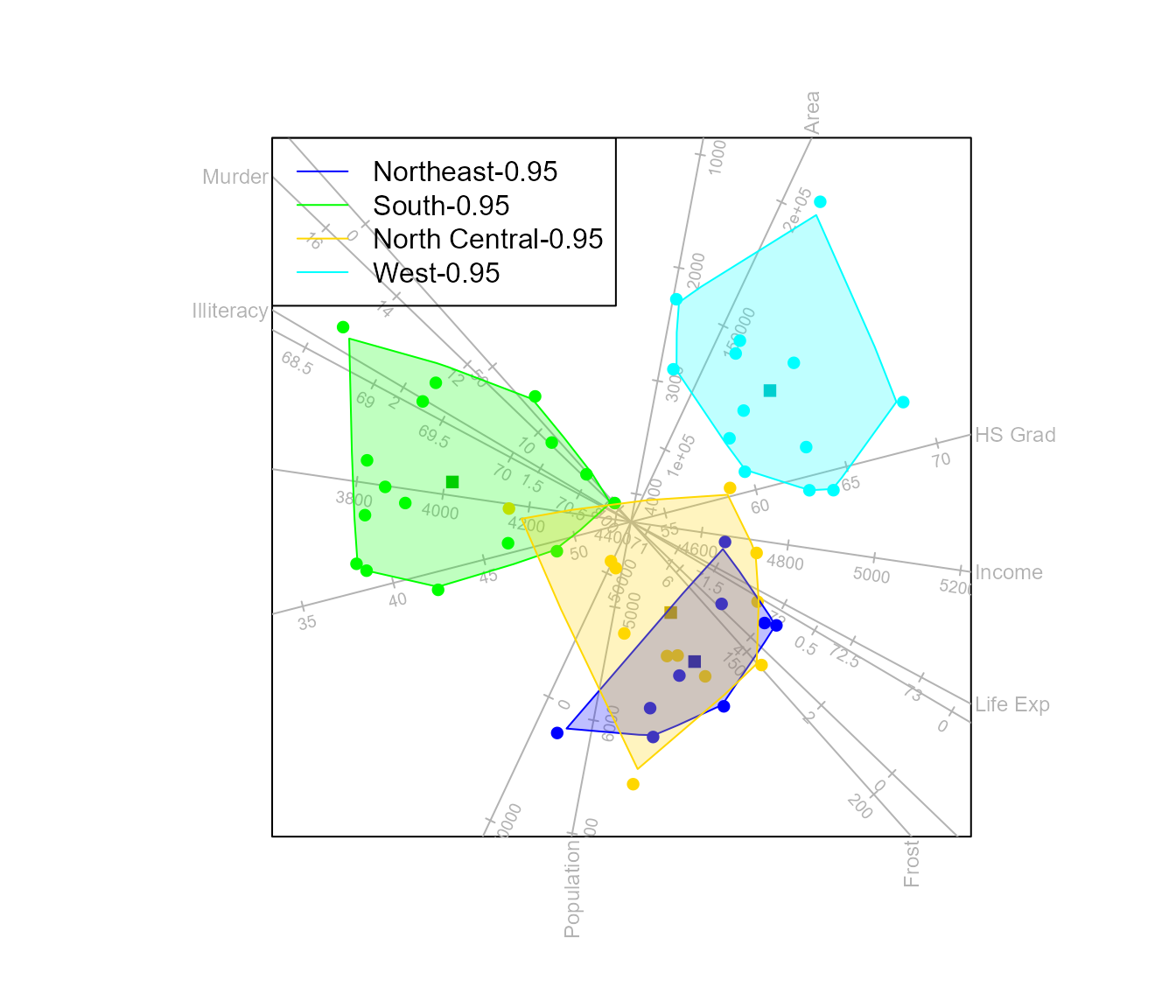
The function means()
This function controls the aesthetics of the class means in the
biplot. The function accepts as first argument an object of class
biplot where the aesthetics should be applied. Let us first
construct a CVA biplot of the state.x77 data with samples
optimally separated according to state.division.
biplot(state.x77, scaled = TRUE) |>
CVA(classes = state.division) |>
legend.type(means = TRUE) |>
plot()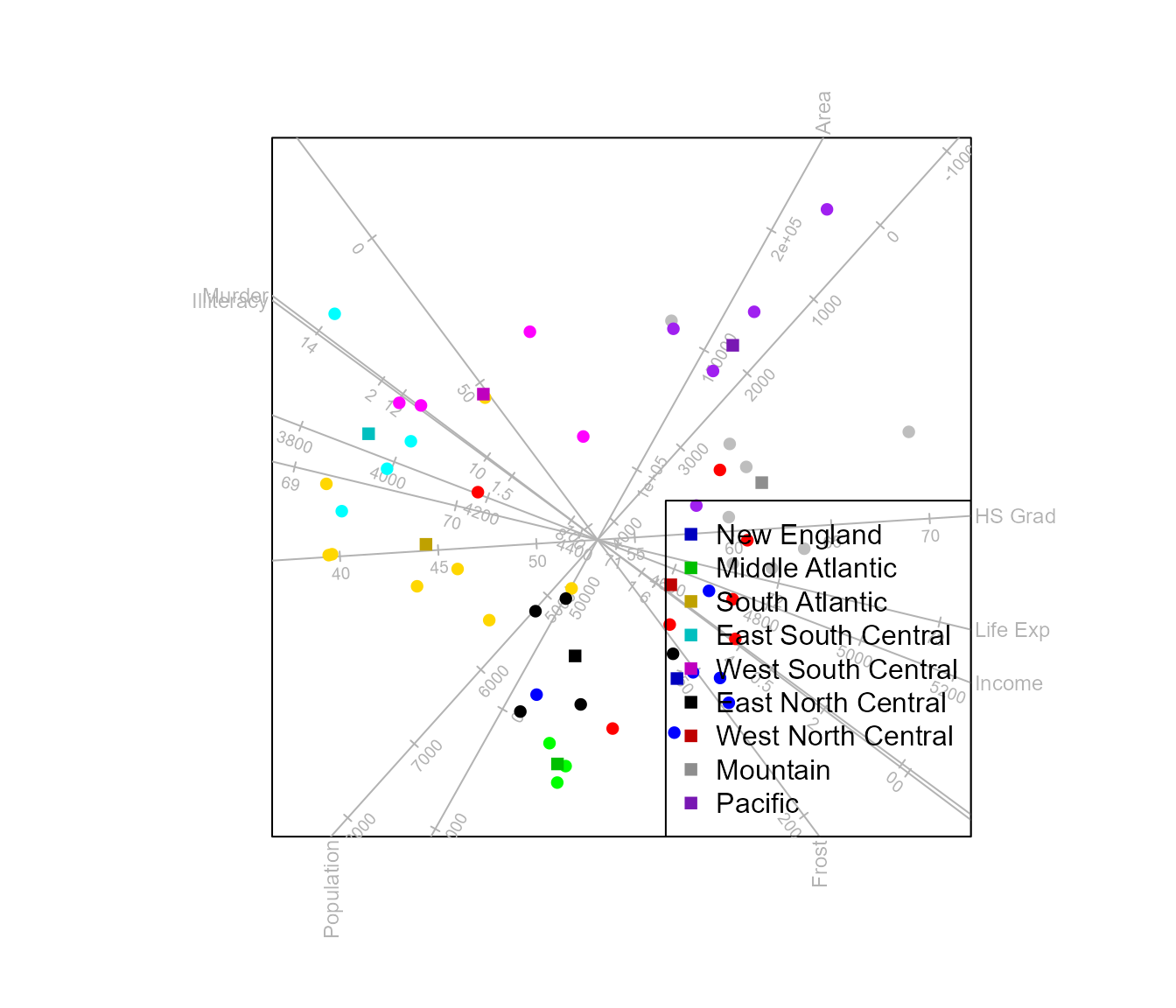
Instead of adding a legend, we can choose to label the class means. Furthermore, the colour of each class mean defaults to the colour of the samples. We wish to select a different colour and plotting character for the class means.
biplot(state.x77, scaled = TRUE) |>
CVA(classes = state.division) |>
means(label = TRUE, col = "olivedrab", pch = 15) |>
plot()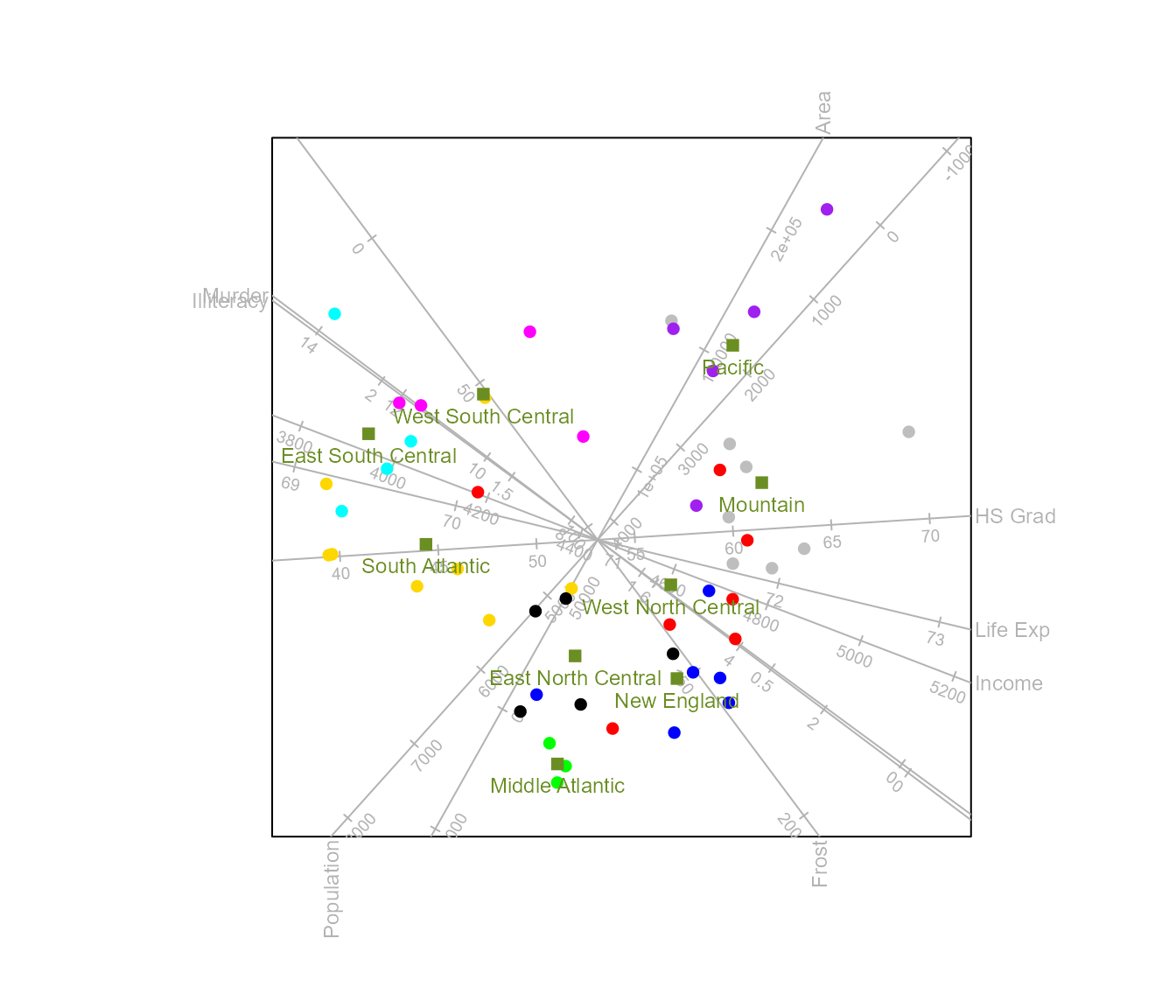
If we choose to only show the class means for the central states, the
argument which is used either indicating the number(s) in
the sequence of levels (which = 4:7), or as shown below,
the levels themselves:
biplot(state.x77, scaled = TRUE) |>
CVA(classes = state.division) |>
means (which = c("West North Central", "West South Central", "East South Central",
"East North Central"), label = TRUE) |>
plot()
The size of the labels is controlled with label.cex
which can be specified either as a single value (for all class means) or
a vector indicating size values for each individual sample. The colour
of the labels defaults to the colour(s) of the class means. However,
individual label colours can be spesified with label.col,
similar to label.cex as either a single value of a vector
of length equal to the number of classes.
biplot(state.x77, scaled = TRUE) |>
CVA(classes = state.division) |>
means (col = "olivedrab", pch = 15, cex = 1.5,
label = TRUE, label.col = c("blue","green","gold","cyan","magenta",
"black","red","grey","purple")) |>
plot()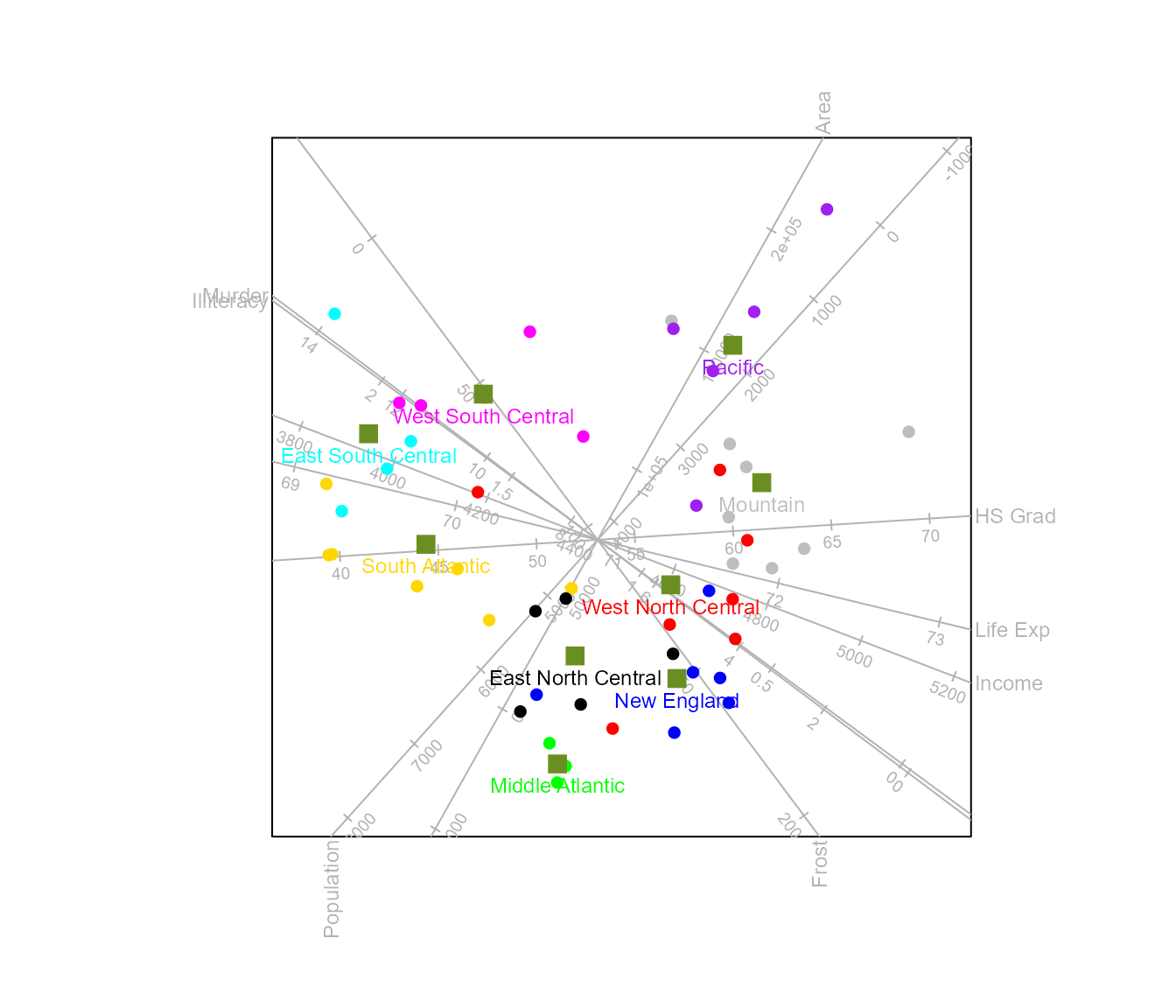
We can also make use of the functionality of the ggrepel
package to place the labels.
biplot(state.x77, scaled = TRUE) |>
CVA(classes = state.division) |>
samples (label = "ggrepel", label.cex=0.65) |>
means (label = "ggrepel", label.cex=0.8) |> plot()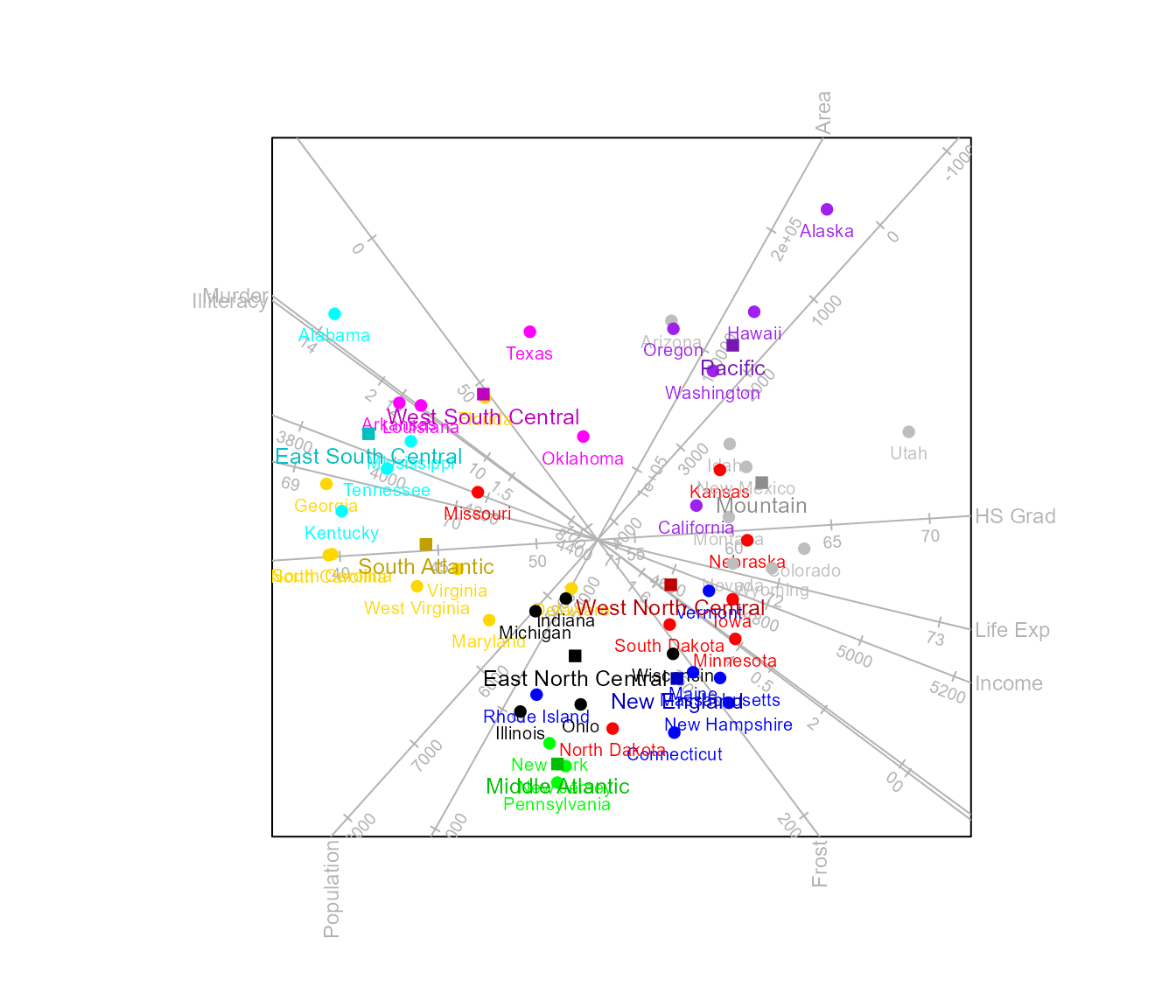
The function classify()
Classification regions can be added to the CVA biplot with the
function classify(). The argument
classify.regions must be set equal to TRUE to
render the regions in the plot. Other arguments such as
col, opacity and borders allows
to change the aesthetics of the regions.
biplot(state.x77, scaled = TRUE) |>
CVA(classes = state.division) |>
classify(classify.regions = TRUE,opacity = 0.2) |>
plot()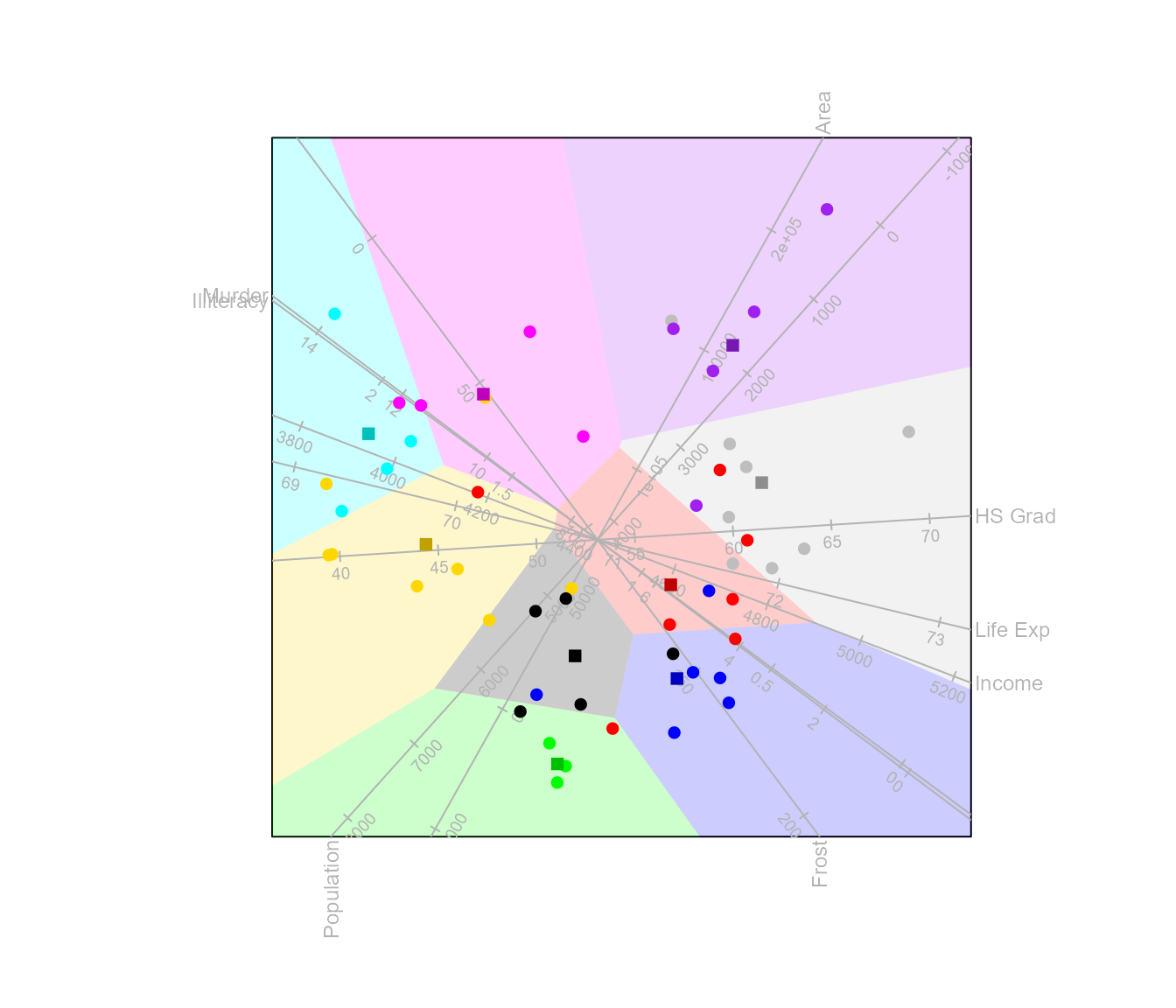
The functions fit.measures() and
summary()
There is a number of fit measures that are specific to CVA biplots.
The measures are computed with the function fit.measures()
and the results are displayed by the function
summary().
Canonical variate analysis can be considered as a transformation of the original variables to the canonical space followed by constructing a PCA biplot of canonical variables. The matrix of class means is transformed to where is a non-singular matrix such that . Pricipal component analysis finds the orthogonal matrix such that
where as defined in section 1. The predicted values for the class means is given by
Overall quality of fit
Based on the two-step process described above, there are two measures of quality of fit. The quality of the approximation of the canonical variables in the -dimensional display is given by
and the quality of the approximation of the original variables in the 2D CVA biplot is given by
Adequacy of representation of variables
The adequacy with which each of the variables is represented in the biplot is given by the elementwise ratios
Predictivity
Between class predictivity
The axis and class mean predictivities are defined in terms of the weighted class means.
Within class predictivity
We define the matrix of samples as deviations from their class means as
where .
Within class axis predictivity
The within class axis predictivity is computed as the elementwise ratios
Within class sample predictivity
Unlike PCA biplots, sample predictivity for CVA biplots are computed for the observations expressed as deviations from their class means. The elementwise ratios is obtained from
To display the fit measures, we create
a biplot object with the measures added by the function
fit.measures() and call summary().
obj <- biplot(state.x77, scaled = TRUE) |>
CVA(classes = state.division) |>
fit.measures() |>
plot()
summary (obj)
#> Object of class biplot, based on 50 samples and 8 variables.
#> 8 numeric variables.
#> 9 classes: New England Middle Atlantic South Atlantic East South Central West South Central East North Central West North Central Mountain Pacific
#>
#> Quality of fit of canonical variables in 2 dimension(s) = 70.7%
#> Quality of fit of original variables in 2 dimension(s) = 70.5%
#> Adequacy of variables in 2 dimension(s):
#> Population Income Illiteracy Life Exp Murder HS Grad Frost
#> 0.41716176 0.15621549 0.16136381 0.09759664 0.19426796 0.55332679 0.50497634
#> Area
#> 0.40661470
#> Axis predictivity in 2 dimension(s):
#> Population Income Illiteracy Life Exp Murder HS Grad Frost
#> 0.1859124 0.4019427 0.8195756 0.6925389 0.7685373 0.9506355 0.7819324
#> Area
#> 0.8458143
#> Class predictivity in 2 dimension(s):
#> New England Middle Atlantic South Atlantic East South Central
#> 0.7922047 0.6570417 0.8191791 0.8777759
#> West South Central East North Central West North Central Mountain
#> 0.7416085 0.6370315 0.3265978 0.6825966
#> Pacific
#> 0.6700194
#> Within class axis predictivity in 2 dimension(s):
#> Population Income Illiteracy Life Exp Murder HS Grad Frost
#> 0.04212318 0.09357501 0.25675620 0.19900223 0.29474972 0.75215233 0.31027358
#> Area
#> 0.12741853
#> Within class sample predictivity in 2 dimension(s):
#> Alabama Alaska Arizona Arkansas California
#> 0.722548912 0.163442379 0.333341120 0.268976273 0.229139828
#> Colorado Connecticut Delaware Florida Georgia
#> 0.264963758 0.082284385 0.593415987 0.461070888 0.636531435
#> Hawaii Idaho Illinois Indiana Iowa
#> 0.015640188 0.113711473 0.338612599 0.389208196 0.507060148
#> Kansas Kentucky Louisiana Maine Maryland
#> 0.784831952 0.314119027 0.078465054 0.008388471 0.306141816
#> Massachusetts Michigan Minnesota Mississippi Missouri
#> 0.076563044 0.218470793 0.645446212 0.046129058 0.710971640
#> Montana Nebraska Nevada New Hampshire New Jersey
#> 0.086279776 0.810374638 0.090490164 0.298187909 0.003496353
#> New Mexico New York North Carolina North Dakota Ohio
#> 0.007134343 0.024268121 0.422776032 0.446240464 0.277262145
#> Oklahoma Oregon Pennsylvania Rhode Island South Carolina
#> 0.450104680 0.108636860 0.033945796 0.415029328 0.261568299
#> South Dakota Tennessee Texas Utah Vermont
#> 0.134881180 0.247921823 0.110537439 0.500454605 0.159941068
#> Virginia Washington West Virginia Wisconsin Wyoming
#> 0.310439564 0.030877305 0.066303623 0.295499472 0.474458397The call to biplot(), CVA() and
fit.measures() is required to (a) create an object of class
biplot, (b) extend the object to class CVA and
(c) compute the fit measures. The call to the function
plot() is optional. It is further possible to select which
fit measures to display in the summary() function where all
measures default to TRUE.
obj <- biplot(state.x77, scaled = TRUE) |>
CVA(classes = state.region) |>
fit.measures()
summary (obj, adequacy = FALSE, within.class.axis.predictivity = FALSE,
within.class.sample.predictivity = FALSE)
#> Object of class biplot, based on 50 samples and 8 variables.
#> 8 numeric variables.
#> 4 classes: Northeast South North Central West
#>
#> Quality of fit of canonical variables in 2 dimension(s) = 91.9%
#> Quality of fit of original variables in 2 dimension(s) = 95.3%
#> Axis predictivity in 2 dimension(s):
#> Population Income Illiteracy Life Exp Murder HS Grad Frost
#> 0.9873763 0.9848608 0.8757913 0.9050208 0.9955088 0.9970346 0.9558192
#> Area
#> 0.9344651
#> Class predictivity in 2 dimension(s):
#> Northeast South North Central West
#> 0.8031465 0.9985089 0.6449906 0.9988469Additional CVA dimensions
It was mentioned that the eigen equation (1) has non-zero eigenvalues. This implies that the CVA biplot for groups, reduces to a single dimension. If we write
the columns of
forms a basis for the orthogonal complement of the canonical space
defined by
_1.
The argument low.dim determines how to uniquely define the
second and third dimensions. By default
low.dim = "sample.opt" which selects the dimensions by
minimising total squared reconstruction error for samples.
The representation of the canonical variates are exact in the first dimension, but not the representation of the individual samples . If we define with a square matrix of zeros except for a in the first diagonal position, then the total square reconstruction error for samples is given by
Define
then is minimised when
where where is the matrix of right singular vectors of .
state.2group <- ifelse(state.division == "New England" |
state.division == "Middle Atlantic" |
state.division == "South Atlantic" |
state.division == "Pacific",
"Coastal", "Central")
biplot (state.x77) |> CVA (state.2group) |> legend.type(means=TRUE) |> plot()
#> Warning in CVA.biplot(biplot(state.x77), state.2group): The dimension of the
#> canonical space < dim.biplot sample.opt method used for additional
#> dimension(s).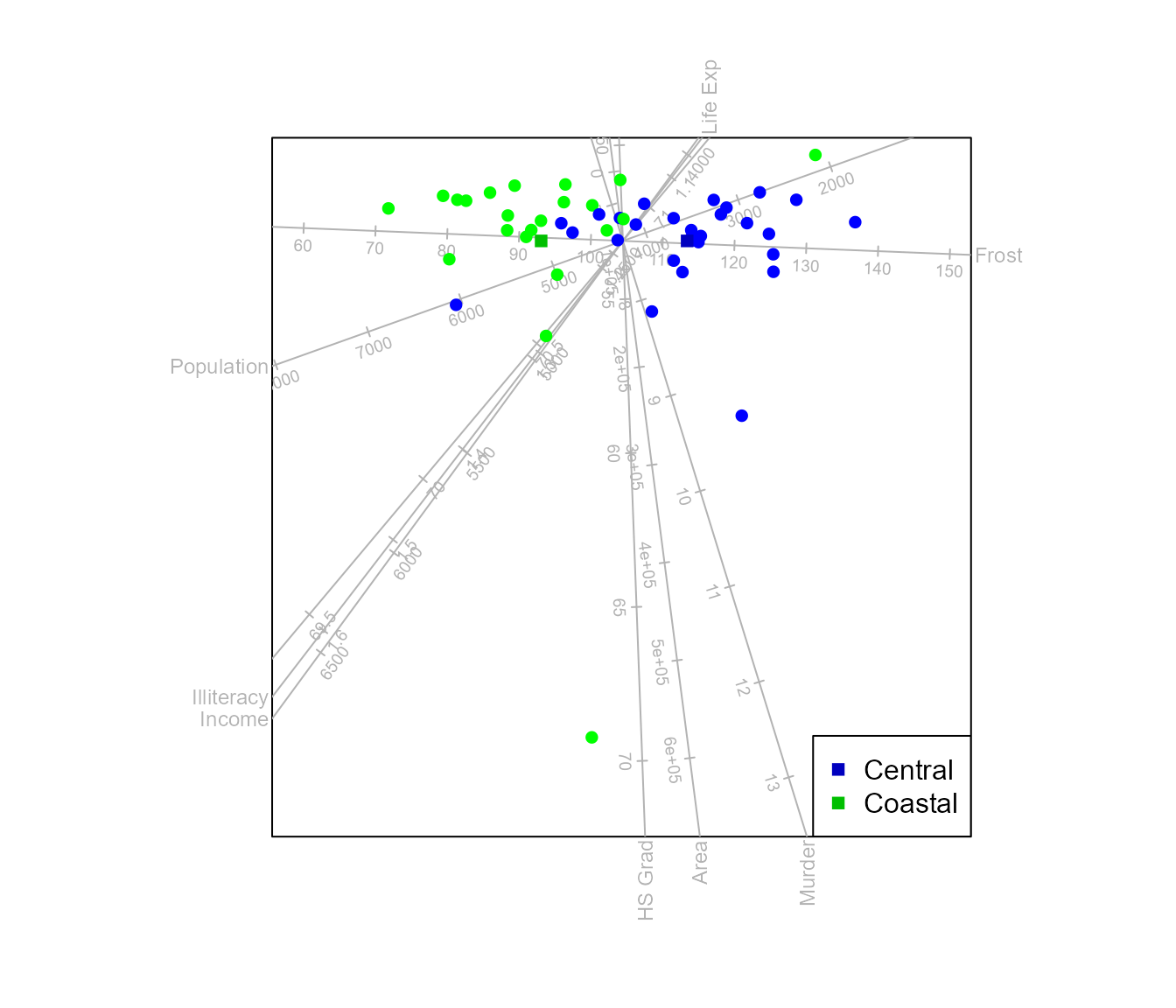
le Roux and Gardner-Lubbe (2024) discuss an alternative method for obtaining additional dimensions. When assuming underlying normal distributions, the Bhattacharyya distance can be optimised. This method is specific to the two class case and cannot be utilised to find a third dimension in a 3D CVA biplot with three classes.
biplot (state.x77) |> CVA (state.2group, low.dim="Bha") |> legend.type(means=TRUE) |> plot()
#> Warning in CVA.biplot(biplot(state.x77), state.2group, low.dim = "Bha"): The
#> dimension of the canonical space < dim.biplot Bhattacharyya.dist method used
#> for additional dimension(s).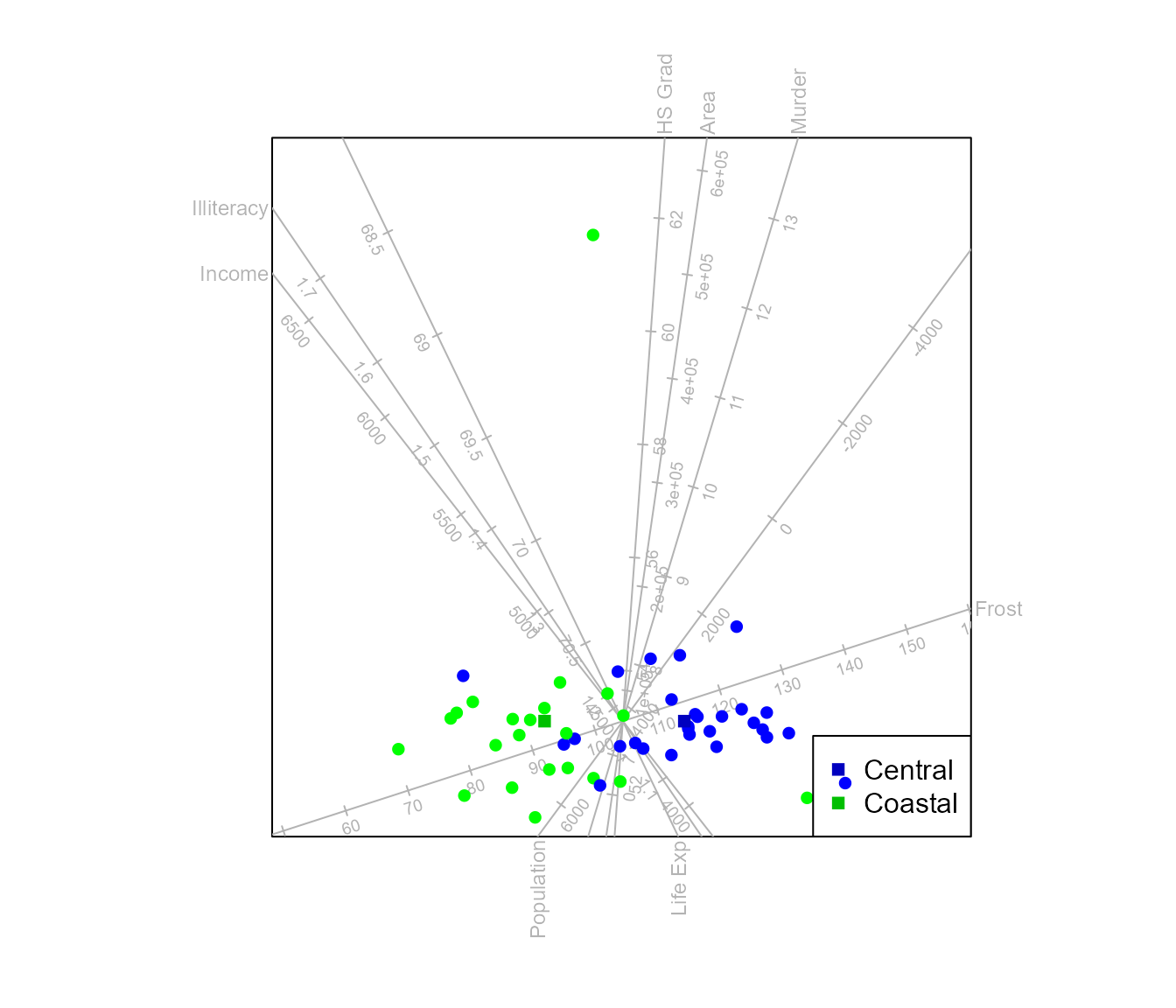
Analysis of Distance (AoD)
Similar to the variance decomposition in CVA, analysis of distance decomposes the total sum of squared distances into a sum of squared distances between class means component and a sum of squared distances within classes component.
Consider any Euclidean embeddable distance metric . For a Euclidean embeddable metric it is possible to find high dimensional coordinates and such that the Euclidean distance between and is equal to . Let the matrix contain the values and similarly the values where represent the distance between class means and .
where . Thus, AoD differs from CVA in allowing any Euclidean embeddable measure of inter-class distance. As with CVA, these distances may be represented in maps with point representing the class means, supplemented by additional points representing the within-group variation. Principal coordinate analysis is performed, only on the matrix .
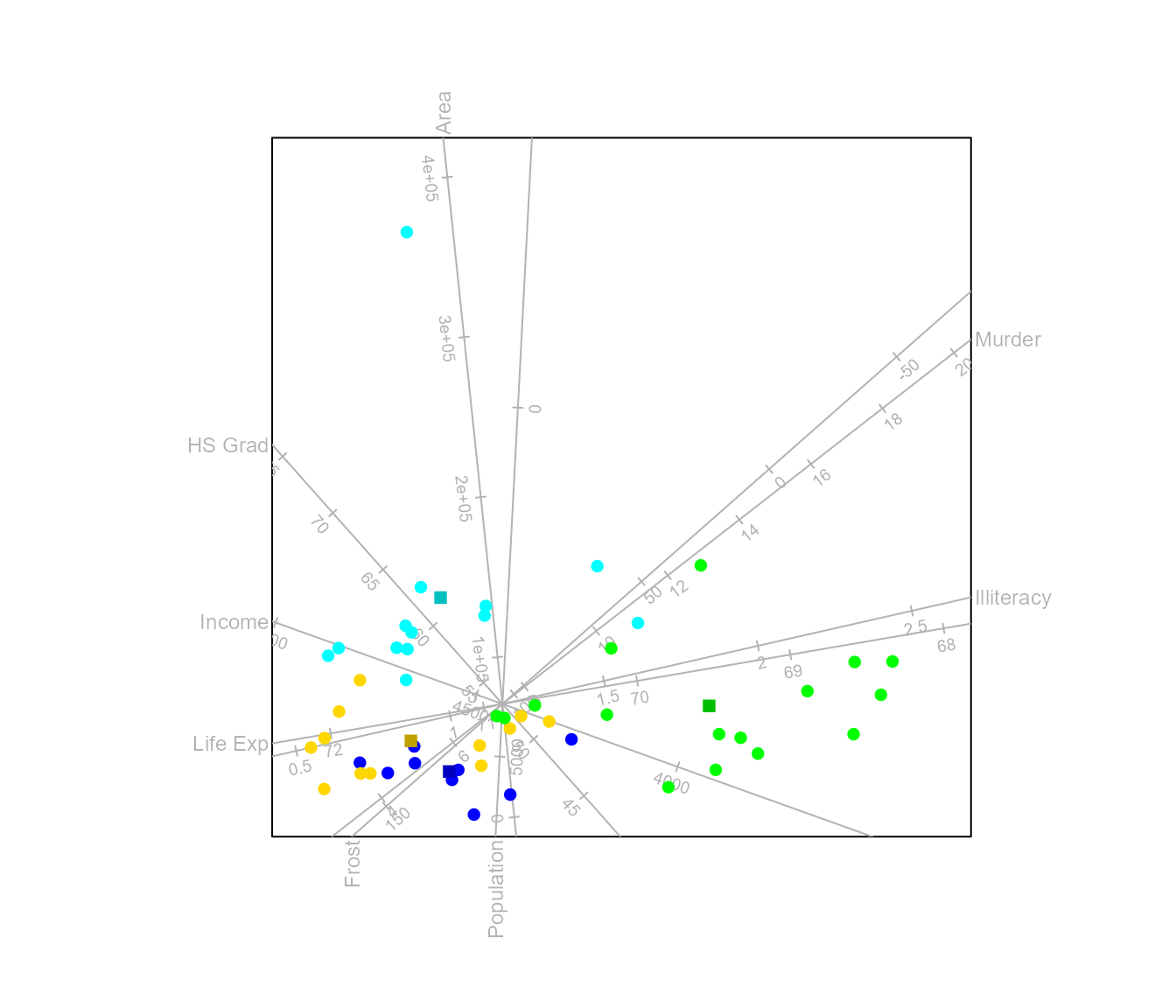
By default linear regression biplot axes are fitted to the plot. Alternatively, spline axes can be constructed.
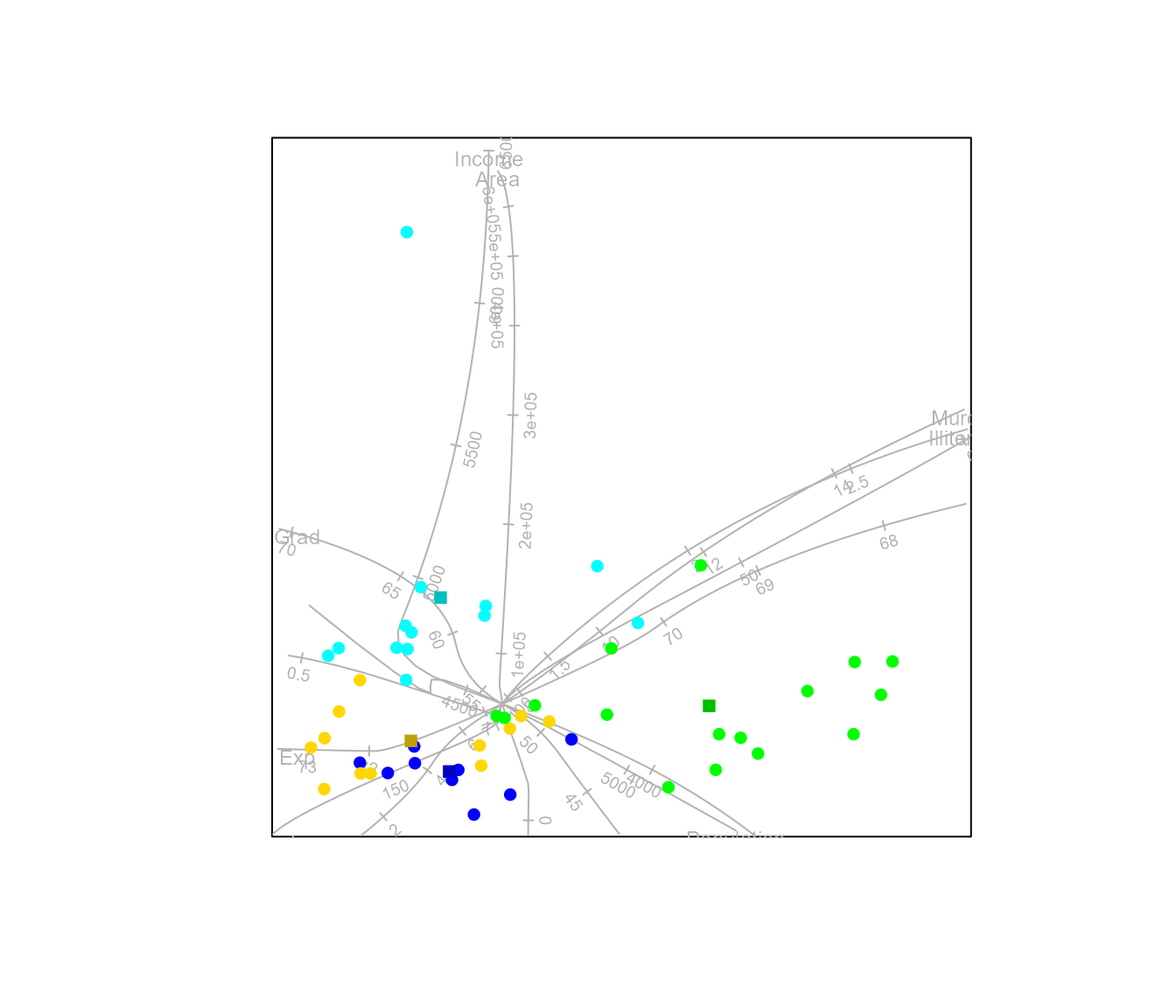
#> Calculating spline axis for variable 1
#> Calculating spline axis for variable 2
#> Calculating spline axis for variable 3
#> Calculating spline axis for variable 4
#> Calculating spline axis for variable 5
#> Calculating spline axis for variable 6
#> Calculating spline axis for variable 7
#> Calculating spline axis for variable 8As an illustration of a Euclidean embeddable distance metric, other than Euclidean distance itself, we can construct an AoD biplot with the square root of the Manhattan distance.
biplot(state.x77, scaled = TRUE) |>
AoD(classes = state.region, axes = "splines", dist.func=sqrtManhattan) |> plot()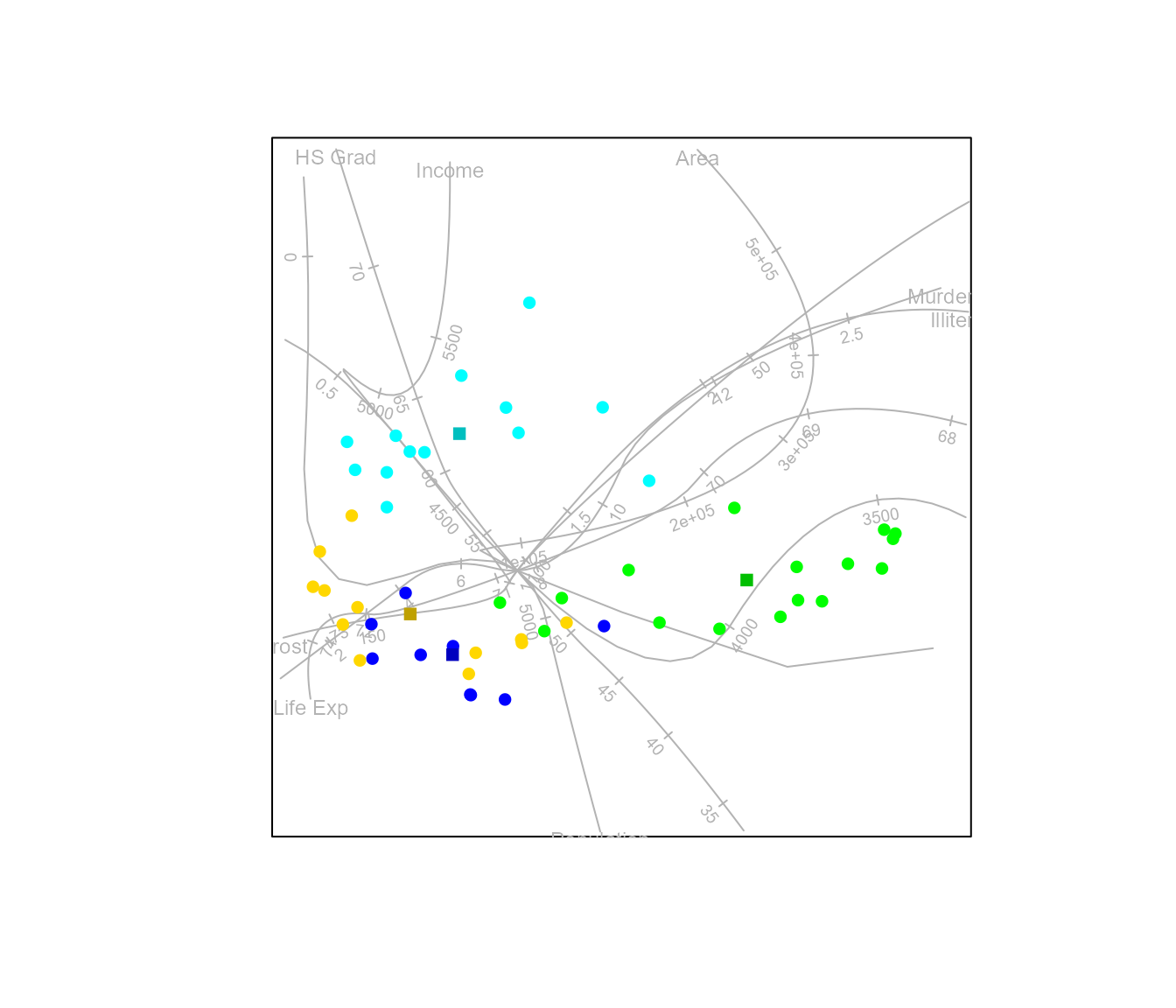
#> Calculating spline axis for variable 1
#> Calculating spline axis for variable 2
#> Calculating spline axis for variable 3
#> Calculating spline axis for variable 4
#> Calculating spline axis for variable 5
#> Calculating spline axis for variable 6
#> Calculating spline axis for variable 7
#> Calculating spline axis for variable 8