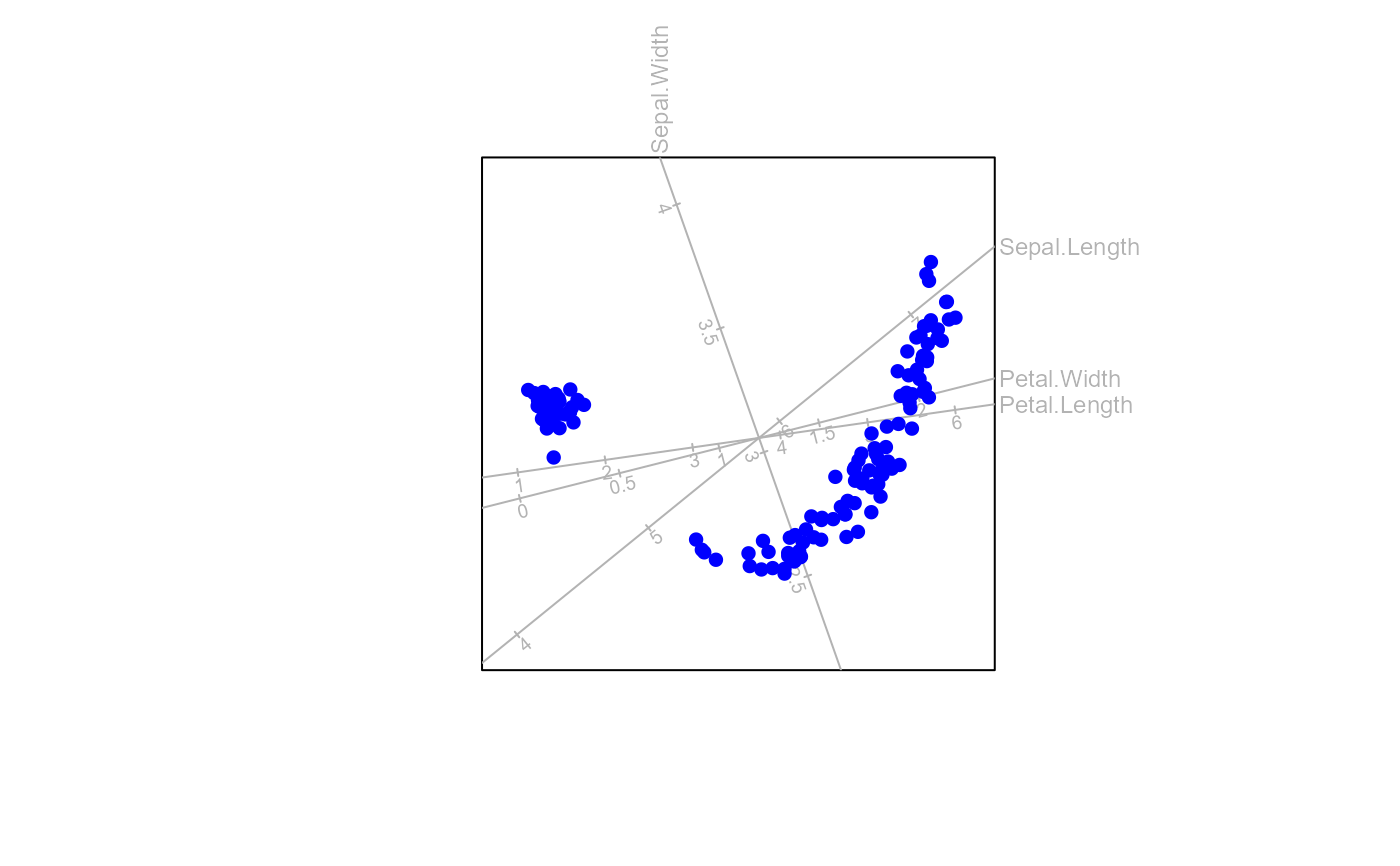
Principal Coordinate Analysis (PCO) biplot method
PCO.RdPrincipal Coordinate Analysis (PCO) biplot method
Arguments
- bp
an object of class
biplotobtained from preceding functionbiplot().- Dmat
nxn matrix of Euclidean embeddable distances between samples
- dist.func
function to compute Euclidean embeddable distances between samples. The default NULL computes Euclidean distance.
- dist.func.cat
function to compute Euclidean embeddable distance between categorical variables for the samples. The default NULL computes the extended matching coefficient.
- dim.biplot
dimension of the biplot. Only values 1, 2 and 3 are accepted, with default
2.- e.vects
e.vects which eigenvectors (canonical variates) to extract, with default
1:dim.biplot.- group.aes
vector of the same length as the number of rows in the data matrix for differentiated aesthetics for samples.
- show.class.means
logical, indicating whether to plot the class means on the biplot.
- axes
type of biplot axes, currently only regression axes are implemented
- ...
more arguments to
dist.func